The raster package for R provides a variety of functions for the analysis of raster GIS data. The focal() function is very useful for applying moving window filters to such data. I wanted to calculate a moving window mean for cells within a specified radius, but focal() did not provide a built-in option for this. The following code generates an appropriate weights matrix for implementing such a filter, by using the matrix as the w argument of focal().
require(raster)
#function to make a circular weights matrix of given radius and resolution
#NB radius must me an even multiple of res!
make_circ_filter<-function(radius, res){
circ_filter<-matrix(NA, nrow=1+(2*radius/res), ncol=1+(2*radius/res))
dimnames(circ_filter)[[1]]<-seq(-radius, radius, by=res)
dimnames(circ_filter)[[2]]<-seq(-radius, radius, by=res)
sweeper<-function(mat){
for(row in 1:nrow(mat)){
for(col in 1:ncol(mat)){
dist<-sqrt((as.numeric(dimnames(mat)[[1]])[row])^2 +
(as.numeric(dimnames(mat)[[1]])[col])^2)
if(dist<=radius) {mat[row, col]<-1}
}
}
return(mat)
}
out<-sweeper(circ_filter)
return(out)
}
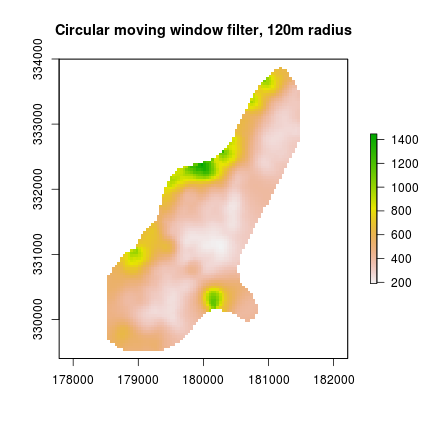
This example uses a weighs matrix generated by make_circ_filter() to compute a circular moving average on the Meuse river grid data. For a small raster like this, the function is more than adequate. For large raster datasets, it’s quite slow though.
#make a circular filter with 120m radius, and 40m resolution
cf<-make_circ_filter(120, 40)
#test it on the meuse grid data
f <- system.file("external/test.grd", package="raster")
r <- raster(f)
r_filt<-focal(r, w=cf, fun=mean, na.rm=T)
plot(r, main="Raw data") #original data
plot(r_filt, main="Circular moving window filter, 120m radius") #filtered data