Readers might recall my earlier efforts at using R and python for geolocation and mapping of realtime fire and emergency incident data provided as rss feeds by the Victorian Country Fire Authority (CFA). My realisation that the CFA’s rss feeds are actually implemented using georss (i.e. they already contain locational data in the form of latitudes and longitudes for each incident), makes my crude implementation of a geolocation process in my earlier python program redundant, if not an interesting learning experience.
I provide here an quick R program for mapping current CFA fire and emergency incidents from the CFA’s georss, using the excellent ggmap package to render the underlying map, with map data from google maps.
Here’s the code:
library(ggmap)
library(XML)
library(reshape)
#download and parse the georss data to obtain the incident locations:
cfaincidents<-xmlInternalTreeParse("http://osom.cfa.vic.gov.au/public/osom/IN_COMING.rss")
cfapoints <- sapply(getNodeSet(cfaincidents, "//georss:point"), xmlValue)
cfacoords<-colsplit(cfapoints, " ", names=c("Latitude", "Longitude"))
#map the incidents onto a google map using ggmap
library(ggmap)
library(XML)
library(reshape)
#download and parse the georss data to obtain the incident locations:
cfaincidents<-xmlInternalTreeParse("http://osom.cfa.vic.gov.au/public/osom/IN_COMING.rss")
cfapoints <- sapply(getNodeSet(cfaincidents, "//georss:point"), xmlValue)
cfacoords<-colsplit(cfapoints, " ", names=c("Latitude", "Longitude"))
#map the incidents onto a google map using ggmap
png("map.png", width=700, height=700)
timestring<-format(Sys.time(), "%d %B %Y, %H:%m" )
titlestring<-paste("Current CFA incidents at", timestring)
map<-get_map(location = "Victoria, Australia", zoom=7, source="google", maptype="terrain")
ggmap(map, extent="device")+
geom_point(data = cfacoords, aes(x = Longitude, y = Latitude), size = 4, pch=17, color="red")+
opts(title=titlestring)
dev.off()
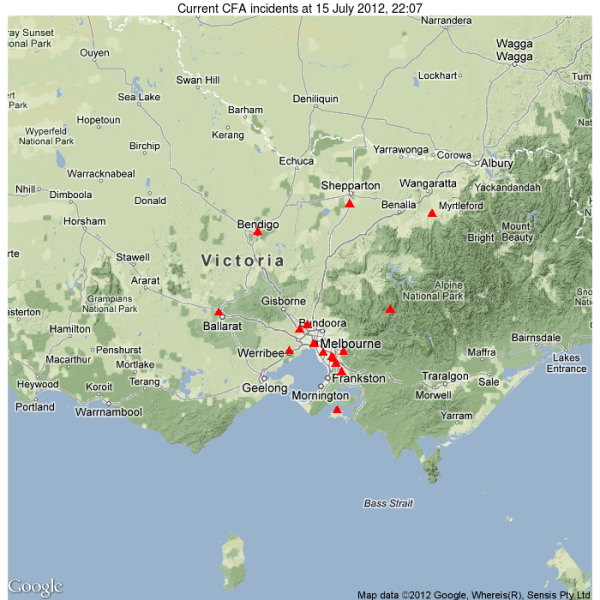
And here’s the resulting map, showing the locations of tonight’s incidents. Note that this is a snapshot of incidents at the time of writing, and should not be assumed to represent the locations of incidents at other times, or used for anything other than your own amusement or edification. The authoritative source of incident data is always the CFAs own website and rss feeds.