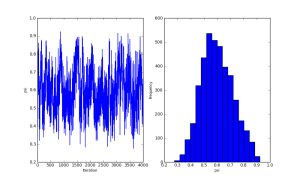
I’ve continued my experimentation with PyMC, using it to fit occupancy models to wildlife survey data with imperfect detectability. Chris Fonnesbeck has provided code for a simple, single-survey occupancy model here, which provides a good starting point for experimentation. I wanted to construct my model using the alternative, state-space parameterisation for occupancy models described by Royle and Kéry(2007). Unlike the multi-season, dynamic occupancy model described by Royle and Kéry, I am only fitting a single-season occupancy model, where the site-states (occupied or unoccupied) are assumed to be constant. The model uses a hierarchical approach, where sites are occupied with probability , and the true occupancy states of the sites,
are inferred from repeated surveys at each site based on a probabilistic detection model (in this case a simple Bernoulli model, with conditional probability of detection at each survey
). Fitting this model using MCMC has the advantage that a finite-sample estimate of occupancy rates among the sampled sites can be easily computed by sampling from
.
from pymc import *
from numpy import *
"""
Alternative implementation of single season occupancy estimation for the Salamander data (from MacKenzie et al. 2006), using a
state-space approach.
Modified from original example code and data provided by Chris Fonnesbeck at https://github.com/pymc-devs/pymc/wiki/Salamanders
"""
# Occupancy data - rows are sites, with replicate surveys conducted at each site
salamanders = array([[0,0,0,1,1], [0,1,0,0,0], [0,1,0,0,0], [1,1,1,1,0], [0,0,1,0,0],
[0,0,1,0,0], [0,0,1,0,0], [0,0,1,0,0], [0,0,1,0,0], [1,0,0,0,0],
[0,0,1,1,1], [0,0,1,1,1], [1,0,0,1,1], [1,0,1,1,0], [0,0,0,0,0],
[0,0,0,0,0], [0,0,0,0,0], [0,0,0,0,0], [0,0,0,0,0], [0,0,0,0,0],
[0,0,0,0,0], [0,0,0,0,0], [0,0,0,0,0], [0,0,0,0,0], [0,0,0,0,0],
[0,0,0,0,0], [0,0,0,0,0], [0,0,0,0,0], [0,0,0,0,0], [0,0,0,0,0],
[0,0,0,0,0], [0,0,0,0,0], [0,0,0,0,0], [0,0,0,0,0], [0,0,0,0,0],
[0,0,0,1,0], [0,0,0,1,0], [0,0,0,0,1], [0,0,0,0,1]])
# Number of replicate surveys at each site
k = 5
#number of detections at each site (row sums of the data)
y=salamanders.sum(axis=1)
#vector of known/unknown occupancies to provide sensible starting values for latent states, z.
#Equal to 1 if at least 1 detection, otherwise zero.
z_start = y>0
# Prior on probability of detection
p = Beta('p', alpha=1, beta=1, value=0.99)
# Prior on probability of occupancy
psi = Beta('psi', alpha=1, beta=1, value=0.01)
#latent states for occupancy
z = Bernoulli('z', p=psi, value=z_start, plot=False)
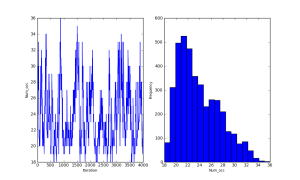
#Number of truly occupied sites in the sample (finite-sample occupancy)
@deterministic(plot=True)
def Num_occ(z=z):
out = sum(z)
return out
#unconditional probabilities of detection at each site (zero for unoccupied sites, p for occupied sites)
@deterministic(plot=False)
def pdet(z=z, p=p):
out = z*p
return out
#likelihood
Y = Binomial('Y', n=k, p=pdet, value=y, observed=True)
Fitting of the model was accomplished by running the following code, which constructs the model, collects some MCMC samples for parameters of interest, and generates plots of the results:
from pylab import * from pymc import * import model #get all the variables from the model file M = MCMC(model) #draw samples M.sample(iter =40000, burn = 20000, thin = 5) #plot results Matplot.plot(M)
Her are some summary plots (traces and histograms) for the MCMC samples of the parameters and
The same model can easily be fitted using OpenBUGS, with comparable results..
model{
psi~dbeta(1, 1)
p~dbeta(1, 1)
for(i in 1:sites){
z[i]~dbern(psi)
pdet[i]<-p*z[i]
for(j in 1:surveys){
Y[i,j]~dbern(pdet[i])
}
}
}
data
list(
surveys=5,
sites=39,
Y=structure(.Data =
c(0,0,0,1,1,
0,1,0,0,0,
0,1,0,0,0,
1,1,1,1,0,
0,0,1,0,0,
0,0,1,0,0,
0,0,1,0,0,
0,0,1,0,0,
0,0,1,0,0,
1,0,0,0,0,
0,0,1,1,1,
0,0,1,1,1,
1,0,0,1,1,
1,0,1,1,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,0,0,
0,0,0,1,0,
0,0,0,1,0,
0,0,0,0,1,
0,0,0,0,1),
.Dim=c(39,5))
)